Project 07:
The role of cotranslational protein biogenesis factors in the translation and targeting of mitochondrial precursor proteins

Christian Münch
E-Mail: ch.muench@em.uni-frankfurt.de
Phone: +49 69-6301-3715
Institute of Molecular Systems Medicine
Goethe University Frankfurt – Medical School

Elke Deuerling
E-Mail: elke.deuerling@uni-konstanz.de
Phone: +49 7531882647
Department of Biology
University of Konstanz
Abstract
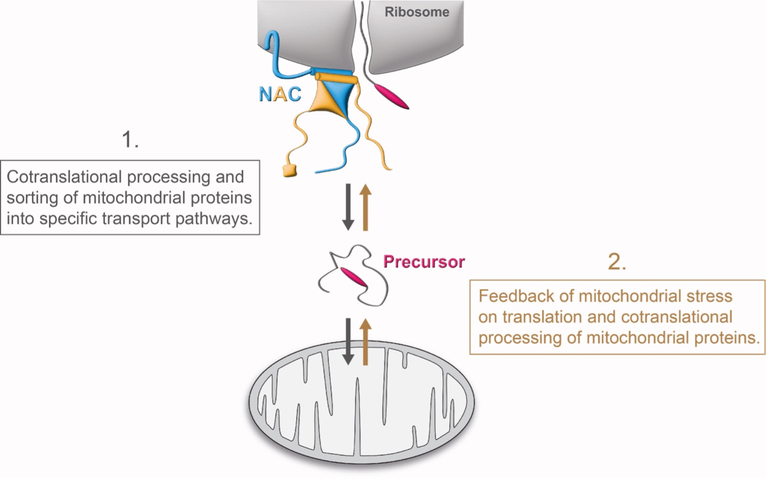
Most mitochondrial proteins are translated in the cytosol and imported into mitochondria. This project will study whether cotranslational processing factors are important for mitochondrial protein biogenesis and transport. In addition, we will investigate feedback loops from mitochondrial proteostasis and import that regulate cytosolic cotranslational protein biogenesis factors.
Project-relevant publications
Gamerdinger M, Jia M, Schloemer R, Rabl L, Jaskolowski M, Khakzar KM, Ulusoy Z, Wallisch A, Jomaa A, Hunaeus G, Scaiola A, Diederichs K, Ban N, Deuerling E (2023) NAC controls cotranslational N-terminal methionine excision in eukaryotes. Science. 380(6651):1238-1243.
Sutandy FXR, Gößner I, Tascher G, Münch C (2023) A cytosolic surveillance mechanism activates the mitochondrial UPR. Nature. 618(7966):849-854.
Jomaa A, Gamerdinger M, Hsieh HH, Wallisch A, Chandrasekaran V, Ulusoy Z, Scaiola A, Hegde RS, Shan SO, Ban N, Deuerling E (2022) Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting. Science. 375(6583):839-844.
Klann K, Münch C (2022) PBLMM: Peptide-based linear mixed models for differential expression analysis of shotgun proteomics data. J Cell Biochem. 123(3):691-696.
Michaelis JB, Brunstein ME, Bozkurt S, Alves L, Wegner M, Kaulich M, Pohl C, Münch C (2022) Protein import motor complex reacts to mitochondrial misfolding by reducing protein import and activating mitophagy. Nat Commun. 13(1):5164.
Schäfer JA, Bozkurt S, Michaelis JB, Klann K, Münch C (2022) Global mitochondrial protein import proteomics reveal distinct regulation by translation and translocation machinery. Mol Cell. 82(2):435-446.e7.
Klann K, Münch C. Instrument logic increases identifications during mutliplexed translatome measurements (2020) Anal Chem. 92(12):8041-8045.
Klann K, Tascher G, Münch C (2020) Functional translatome proteomics reveal converging and dose-dependent regulation by mTORC1 and eIF2α. Mol Cell. 77(4):913-925.e4.
Gamerdinger M, Kobayashi K, Wallisch A, Kreft SG, Sailer C, Schlömer R, Sachs N, Jomaa A, Stengel F, Ban N, Deuerling E (2019) Early scanning of nascent polypeptides inside the ribosomal tunnel by NAC. Mol Cell. 75(5):996-1006.e8.
Shen K, Gamerdinger M, Chan R, Gense K, Martin EM, Sachs N, Knight PD, Schlömer R, Calabrese AN, Stewart KL, Leiendecker L, Baghel A, Radford SE, Frydman J, Deuerling E (2019) Dual role of ribosome-binding domain of NAC as a potent suppressor of protein aggregation and aging-related proteinopathies. Mol Cell. 74(4):729-741.e7.
Read more
All Projects
Find out more about the other tandem/tridem projects that are conducted as part of the SPP2453.
Scientific Background
See what is already known in the mitostasis research area and get to know the central questions of the SPP2453.
Structure
See how the Priority Program SPP2453 is organized and whom to contact if you have questions.